News
Witnessing the dance of RNAs around droplets of cellular degradation machinery
By Mandira Banerjee | 17 April | news.umich.edu
If a picture's worth a thousand words, a video is perhaps worth much more.
For the first time, scientists have observed a dynamic localization of a single RNA with processing bodies—the membraneless organelles present in the cytoplasm of nearly all mammalian cells that play an important role in RNA regulation. RNAs are essential for the normal functioning of a cell.
Until now, scientists have only looked at static snapshots of several RNAs localized at processing bodies, or p-bodies, obscuring a delicate dance. By employing a cutting-edge, two-color fluorescence microscopy technique, the researchers were able to study the real-time kinetic mechanisms of RNAs in live cells at the single-molecule level.
 "Whether an RNA is static or dynamic at the p-body depends on the RNA's size and function," said Sethuramasundaram Pitchiaya, lead author of the study and research investigator at the Michigan Center for Translational Pathology in the U-M Medical School. "We also found that p-bodies are not designated cellular sites for RNA degradation. RNAs can get degraded either inside or outside p-bodies."
"Whether an RNA is static or dynamic at the p-body depends on the RNA's size and function," said Sethuramasundaram Pitchiaya, lead author of the study and research investigator at the Michigan Center for Translational Pathology in the U-M Medical School. "We also found that p-bodies are not designated cellular sites for RNA degradation. RNAs can get degraded either inside or outside p-bodies."
There are a plethora of RNAs in our bodies, which dominate the expression patterns of all genes in all cells. The p-bodies, which behave similarly to oil droplets in water, are enriched with RNA-binding proteins and decay factors and are believed to be cellular sites of RNA degradation.
Lead researcher Nils Walter says the finding has immense potential for drug screening and targeting diseased cells.
"If one can shuttle microRNAs or messenger RNA (mRNA) specifically to p-bodies, unwanted RNAs can be removed from diseased cells. Conversely, the needed RNAs can potentially be recovered from p-bodies to be expressed again," said Walter, the Francis S. Collins Collegiate Professor of Chemistry, Biophysics and Biological Chemistry.
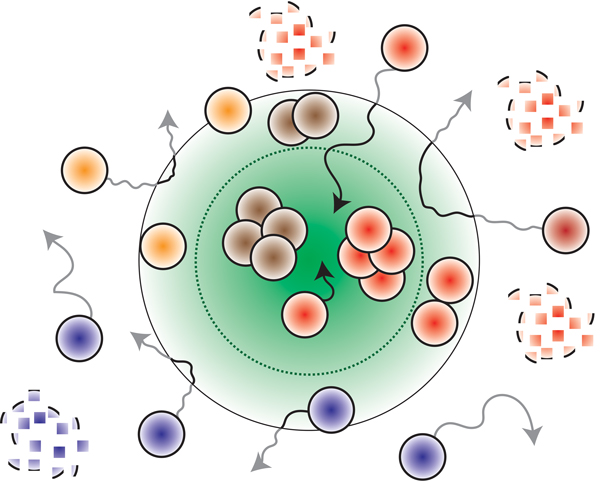 What was unknown before this study was whether all RNAs irrespective of their size and function can assemble into p-bodies and whether the trapped RNAs are degraded at these sites.
What was unknown before this study was whether all RNAs irrespective of their size and function can assemble into p-bodies and whether the trapped RNAs are degraded at these sites.
The new research shows that RNAs are not merely stuck in p-bodies, but can dynamically associate with p-bodies—moving rapidly within p-bodies, getting recruited from the cytoplasm into p-bodies or exiting from p-bodies into the cytoplasm.
"In addition to storing repressed mRNA, we now know that the p-body can also degrade the unused microRNAs for their surveillance," said Santiago Schnell, professor of molecular & integrative physiology.
According to the researchers, the data from the study suggest that the fundamental principles of physical chemistry hold true for mRNA-regulation processes within the complex cellular environment.
The insights from this finding can be applied toward gene therapy and antiviral and antibiotic drugs that target pathogenic RNA function.
Other authors include Arul Chinnaiyan from the U-M Rogel Cancer Center, Marcio Mourao of the U-M Department of Molecular & Integrative Physiology, Ameya Jalihal of the U-M Department of Chemistry, and Lanbo Xiao and Xia Jiang of the Michigan Center for Translational Pathology.
The study is published in the journal Molecular Cell.
