Our Research
The DiFeo lab strives to make original and impactful contributions to the field of women’s cancer research. Dr. DiFeo’s broad background in cancer genetics and biology and specific expertise in miRNA biology, chemotherapy resistance, tumor initiation, and development of patient-based models have positioned her laboratory to work on research projects that span the translational research continuum. Beginning with an in-depth analysis of patient tumors, then progressing to a functional assessment of key genetic drivers of ovarian cancer progression, and ultimately to the development of novel therapeutic approaches to abrogate these drivers to uncover therapies that will improve ovarian cancer patient survival. To accomplish this, we focus on three major research areas: 1) the generation of clinically relevant gynecological cancer models, 2) the discovery of potent genetic drivers with a focus on microRNAs involved in tumor initiation, drug resistance, and recurrence, and 3) development of novel or re-purposed drugs.
RESEARCH AREAS
Generation of Clinically Relevant Gynecological Cancer Models
Since the inception of the lab, we have devoted a substantial amount of time to fostering collaborations with clinicians to build a well-integrated research program that would promote patient-focused, impactful research ideas and permit the development of a large repository of gynecologic tumors and patient models. Through various gynecologic cancer tumor repositories across New York, Cleveland, and Michigan the DiFeo laboratory has been able to collect over 500 advanced-stage uterine and ovarian tumors. Most importantly, we have developed and fully characterized numerous patient-derived cell lines, organoids, and xenografts (PDX) which conserve original tumor characteristics such as heterogeneous histology, clinical biomolecular signature, malignant phenotypes, and genotypes. Given that several studies have shown that many of the commercially available ovarian cancer cell lines do not recapitulate the genomic signature of patient tumors this resource has been used by investigators across the country. We have shared these models with numerous collaborators in the scientific community as well as licensed these models to various CROs, which has led to the discovery of novel therapeutic targets, biomarkers, patent applications, and drugs.1-20 In addition, drug companies including Eli Lilly, Immunogen, SPARC/Sun Pharma, and Benevolent AI have collaborated on sponsored studies with the DiFeo laboratory further highlighting the distinctive feature of this resource.
Identifying Functional Drivers Involved in Ovarian Cancer Progression
The DiFeo played a central role in defining KLF6 and its novel oncogenic splice variant, KLF6-SV1, as a key regulator of several human malignancies.21-34 Our findings were the first to demonstrate that the large majority of ovarian cancer tumors have increased expression of the oncogenic KLF6-SV1 isoform and high levels of this protein correlated with increased tumor aggressiveness. Furthermore, through these studies, we discovered that KLF6-SV1 was a novel anti-apoptotic protein that targets the pro-survival molecule NOXA for degradation and its targeted inhibition extends survival ovarian cancer. These studies resulted in 18 publications during my doctoral and early post-doctoral studies in journals such as Science Translational Medicine, Journal of Clinical Investigation, Cancer Research, Clinical Cancer Research, and Oncogene. 21-34
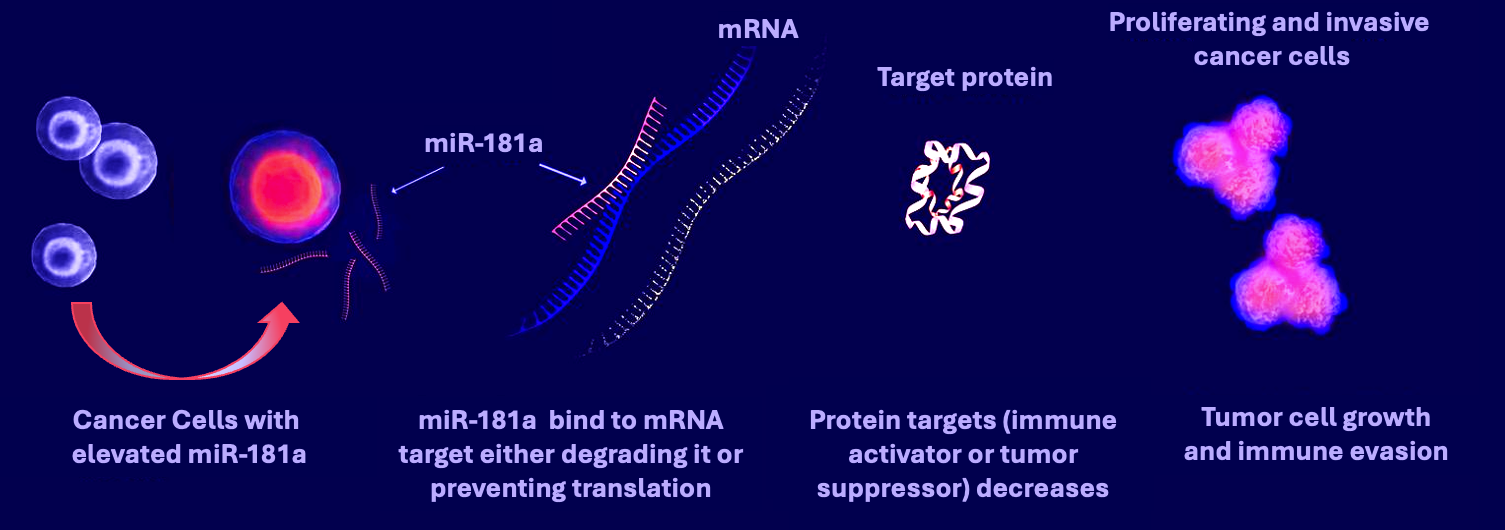
Currently, the DiFeo laboratory is focused on uncovering functional microRNA:mRNA pathways driving ovarian cancer chemotherapy resistance and disease recurrence. We uncovered that microRNA-181a is an oncoMIR wherein it is frequently overexpressed in recurrent, platinum-resistant high-grade serous ovarian cancer (HGSC) and correlates with shorter time to recurrence and poor overall survival. 35 Mechanistically, miR181a contributes to cellular transformation, metastasis initiation, and tumor recurrence by modulating chromosomal instability, EMT, and stemness factors via direct regulation of STING, TGFβ, and Wnt2-13. Most recently, we have shown that in both immunodeficient and immunocompetent HGSC mouse models, targeted inhibition miR181a resulted in decreased tumor dissemination, ascites accumulation, and increased sensitivity to immunotherapy. These studies have resulted in numerous publications including two in Nature Communications. In addition, this research provides the rationale for determining whether pharmacological inhibition of miR181a can simultaneously inhibit the key signaling pathways implicated in cancer progression and significantly enhance patient survival. In addition, several news outlets, including a blog published by the National Cancer Institute, have highlighted the research upon which this application is based. https://www.cancer.gov/news-events/cancer-currents-blog/2020/ovarian-cancer-form-microrna?cid=eb_govdel Lastly, this work has resulted in the development of novel tools, such as a miR181a biosensor and a miR181a transgenic mouse, thereby laying the groundwork for future scientific endeavors and resources.
Development of Novel or Re-purposed Drugs to Treat Platinum-Resistant Cancer
The development of resistance to first-line chemotherapeutics most notably platinum-based therapies leaves few options for the clinical management of advanced ovarian cancer. Therefore, circumventing tumor resistance to commonly used first-line agents represents a very important aspect of multiple initiatives to eliminate ovarian cancer. Thus, another focus of the DiFeo laboratory is to uncover the underlying mechanism of platinum resistance and to develop therapeutic strategies to treat recurrent, chemotherapy-resistant diseases. Through the development of numerous platinum-resistant isogenic primary cell lines as well as patient-derived xenograft models we have uncovered several targetable pathways that can sensitize cells to platinum-based therapies.1-6,8,10,11,14,17-20, 36-39 For example, we are the first to show that altered glutamine metabolism contributes to platinum-resistant ovarian cancer and that targeting glutamine metabolism together with platinum-based chemotherapy offers a potential treatment strategy, particularly in drug-resistant ovarian cancer.4 Our lab has also used various drug repositioning platforms such as “DrugPredict” to identify FDA approved drugs for repositioning as ovarian cancer therapeutics. Using the DrugPredict algorithm, we have identified the Class III antiarrhythmic agent Amiodarone as a potential anti-cancer drug for ovarian cancer treatment6. These findings have allowed us to developed a novel anti-mitotic compound that decouples the potent calcium/potassium channel-blocking activities of this drug but retains the anti-tumorigenic effects.
Lastly, we have recently found that small molecule-mediated stabilization of protein phosphatase 2A (PP2A) modulates the Homologous Recombination pathway and potentiates DNA damage-induced cell death.20 These studies led to the discovery that PP2A modulators can surmount PARP inhibitor insensitivity, and that the combination of PP2A modulators and PARP inhibitors is a novel therapeutic approach for treating recurrent ovarian cancer.
—
REFERENCES