Oct 2020
Combination of menin and FLT3 inhibitors is very effective in AML models. Read in Blood. Featured on the cover!
Aug 2020
We published first in class NSD1 inhibitors in Nature Chemical Biology. Highlighted in Nature Reviews in Drug Discovery.
July 2020
Here is our review on epigenetic protein-protein interactions with Brian as first author
Jan 2020
We reported MI-3454, a very potent menin inhibitor. See our JCI paper.
# news not updated
Jan 2016
We published paper "Property focused structure-based optimization of small molecule inhibitors of the protein-protein interaction between menin and Mixed Lineage Leukemia (MLL)" in the Journal of Medicinal Chemistry.
Nov 2015
We determined structure of ZMIZ1 TPR domain and published paper in collaboration with Mark Chiang: The PIAS-like Coactivator Zmiz1 Is a Direct and Selective Cofactor of Notch1 in T Cell Development and Leukemia. Published in Immunity.
Aug 2015
Our manuscript "Two loops undergoing concerted dynamics regulate activity of the ASH1L histone methyltransferase" has been published in Biochemistry.
Our study "Rational design of orthogonal multipolar interactions with fluorine in protein-ligand complexes" has been published in Journal of Medicinal Chemistry, selected as Editor's choice.
July 2015
Felicia Gray defended her PhD thesis: "Dissecting Bmi1 protein-protein interactions through chemical biology". Congratulations Felicia!
June 2015
We have published study demonstrating that menin-MLL inhibitors block activity of MLL fusion proteins in a mechanism independent on fusion partner. Published in Leukemia.
May 2015
George Lund defended his PhD dissertation: "Targeting CDC25B-CDK2/CyclinA Activity Using Chemical Biology Approaches".
Congratulations George!
April 2015
Our menin-MLL inhibitor program has been featured as cover story in BioCentury Innovations
Dr. Chinnaiyan's lab discovered role of menin and demonstrated efficacy of menin inhibitors in castrate resistant prostate cancer. Study published in Nature Medicine.
March 2015
Our team work resulted in development of potent inhibitors of menin-MLL interaction with strong efficacy in animal models of leukemia. Published in Cancer Cell.
We have licensed menin-MLL inhibitors to Kura Oncology for further development.
Feb 2015
Jola is co-author on study to develop small molecule inhibitors of CBFB-SMMHC: "A small-molecule inhibitor of the aberrant transcription factor CBFβ-SMMHC delays leukemia in mice" published in Science.
Jan 2015
We published review Targeting protein–protein interactions in hematologic malignancies: still a challenge or a great opportunity for future therapies? in Immunological Reviews
Dec 2014
Our study Inhibition of CDC25B Phosphatase Through Disruption of Protein-Protein Interaction has been published in ACS Chemical Biology.
Nov 17, 2014
Jola has been elected to the University of Michigan Medical School "League of Research Excellence". Congratulations to Jola!
Oct 2014
Our study "The same site on LEDGF IBD domain represents therapeutic target for MLL leukemia and HIV" has been published in Blood.
Apr 21, 2014
George's publication "Solution NMR studies reveal no global flexibility in the catalytic domain of CDC25B" has been published in the journal Proteins.
Congratulations to George!
Our review "Challenges and opportunities in targeting the menin-MLL interaction" has been published in Future Medicinal Chemistry
1. Targeting menin-MLL interaction in leukemia
Menin functions as a critical oncogenic cofactor of mixed lineage leukemia (MLL) fusion proteins in the development of acute leukemias. Inhibition of the menin-MLL interactions represents a very promising strategy in reversing oncogenic activity of MLL fusion proteins in acute leukemia.
In our laboratory we determined the first three dimensional structure of menin and employed HTS to identify the first small molecule inhibitors of the menin-MLL protein-protein interaction. Through extensive medicinal chemistry and structure-based design we developed nanomolar menin-MLL inhibitors.
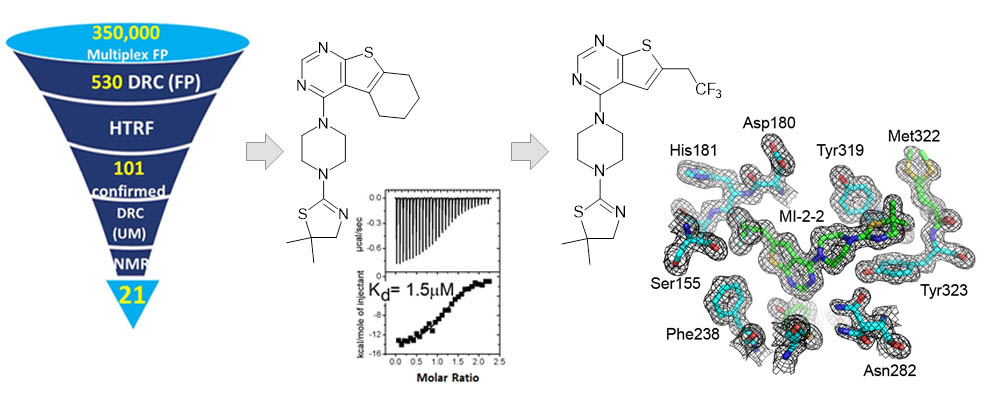
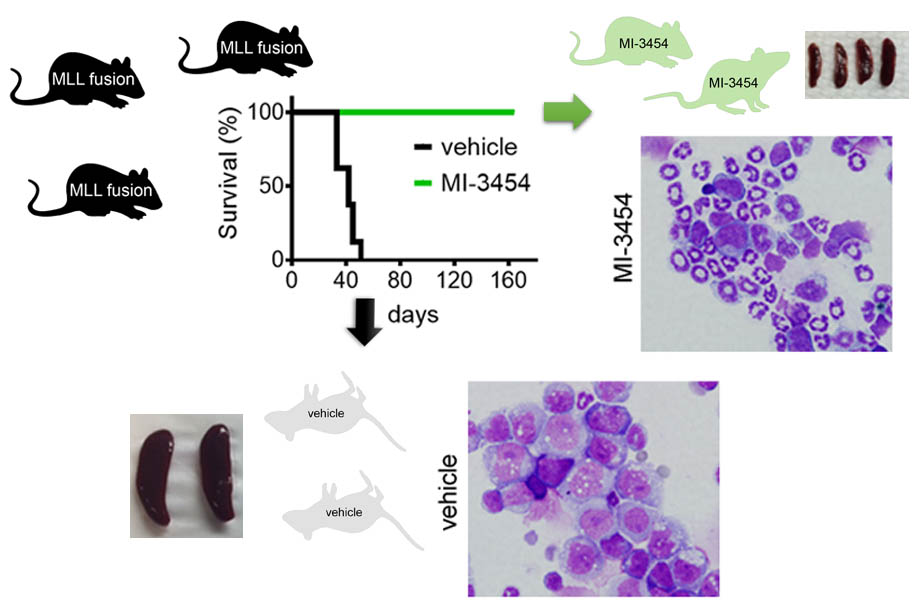
Recently, we developed very potent, sub-nanomolar menin inhibitor MI-3454, with very potent activity in MLL-rearranged and NPM1 mutated leukemia cells and outstanding efficacy in animal models of leukemia.
In collaboration with KURA Oncology we co-developed KO-539, which is currently investigated in phase 1/2a clinical studies.
History and milestones in menin-MLL project
- 2010 Biochemical characterization of menin-MLL interaction (J. Biol. Chem.)
- 2011 Report of the first crystal structure of menin (J. Biol. Chem.)
- 2012 Discovery of the first small molecule inhibitors of menin (Nat. Chem. Biol.)
- 2012 Crystal structure of human menin with bound inhibitor (Blood)
- 2015 Development of MI-503 with in vivo efficacy (Cancer Cell)
- 2019 KURA Oncology starts phase 1/2a studies with KO-539
- 2020 MI-3454 –menin inhibitor with outstanding efficacy in leukemia models (J. Clin. Invest.)
- 2020 Combination of MI-3454 with FLT3 inhibitors results in complete remissions in AML models (Blood)
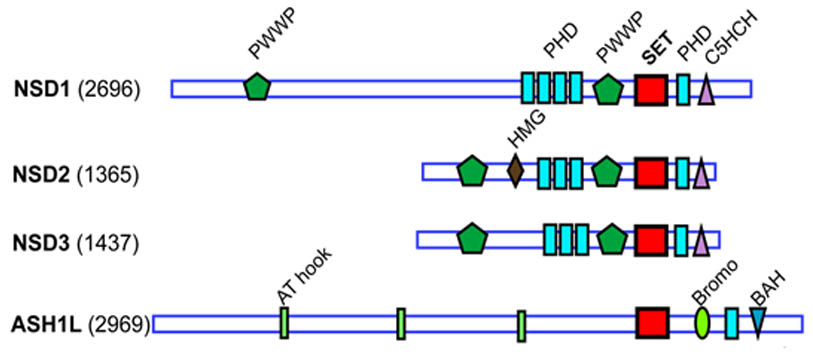
2. Targeting histone methyltransferases
Histone methyltransferases involved in H3K36 methylation are involved in cancers and constitute attractive targets for inhibitor development (read our review on that topic).
NSD1 in NUP98-NSD1 leukemia
Translocations of NUP98-NSD1 are found in aggrieve cases of pediatric leukemias. Past research found that histone-methyltransferase activity of NSD1 SET domain is required for oncogenic activity of NUP98-NSD1. To identify compounds binding to NSD1 catalytic SET domain we employed NMR-based fragment screening approach. By screening our proprietary library of fragments we identified compound BT1.
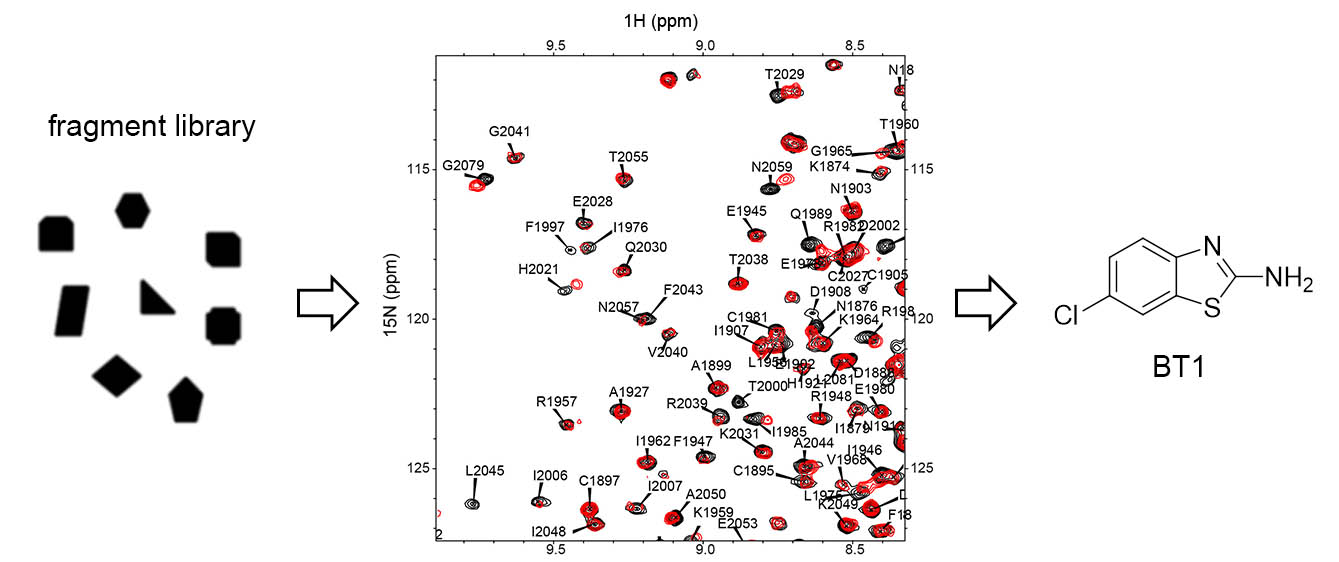
Employing extensive medicinal chemistry we developed compound BT3 that binds covalently to NSD1 and determined the crystal structure of the complex with NSD1. This structure revealed extensive conformational changes in SET domain and channel-like pocket where BT3 binds.
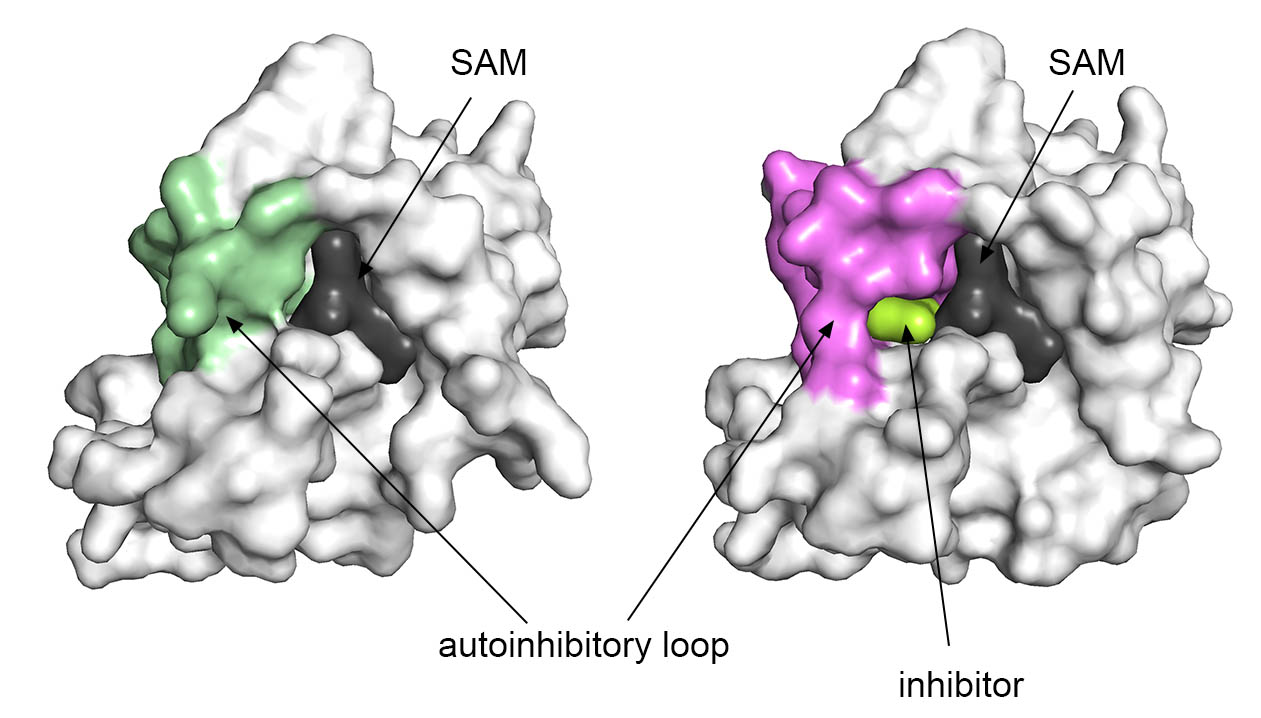
Further medicinal chemistry resulted in development of BT5 - a covalent NSD1 inhibitor. BT5 inhibits NSD1 in cancer cells and block proliferation of leukemia cells with NUP98-NSD1 translocations.
This story has been published in Nature Chemical Biology.
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
 ON THE COVER
ON THE COVER
