Mechanisms of Gene Regulation and Chromatin RemodelingThis project entails elements of other projects described here but focuses on more basic mechanisms of how transcription factors that specify cell lineages and tissues can alter the chromatin at target loci such that epigenetic information is passed on to all progeny cells within the lineage. Again our model protein is the Pax2 transcription factor. We exam how Pax2 is regulated in vivo and in vitro. Using both transgenic mice and novel protein purification methods, we have shown that early activation of the Pax2 intermediate mesoderm specific enhancer requires binding of the chromatin remodeling protein YY1
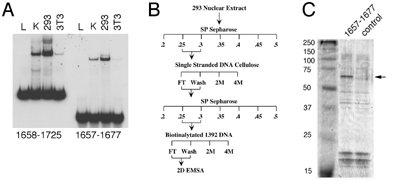
Figure 6 Isolation of the YY1 protein using a combination of biochemical purification, DNA affinity chromatography, and mass spectrometry.
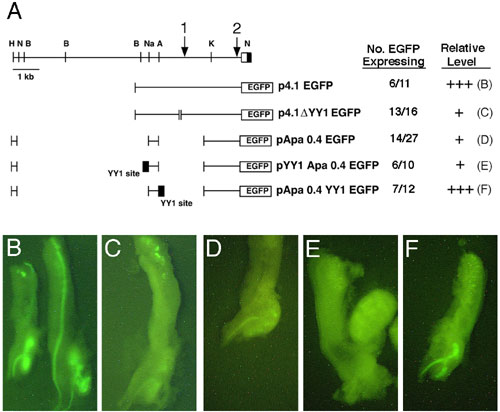
Figure 7 Analysis of the Pax2 promoter and enhancer sequences in transgenic mice using EGFP as a reporter system. Various deletion constructs point to an essential role for the YY1 site in regulating Pax2 expression
One a related note we are studying the interactions of Pax2 with the cellular transcription machinery. Using biochemical approaches, we have identified a Pax2 interacting protein called PTIP that is an essential factor for early development. PTIP interacts with multiple cellular proteins that are involved in chromatin remodeling, DNA damage repair and cell cycle control. We have used protein purification methods and mass spectrometry to identify novel proteins within the Pax2/PTIP complex. Some of these proteins are involved in the modification of Histones, suggesting a link between cell lineage specification and the formation of chromatin domains within specific Pax2 target loci. We are using both classical differential screening methods and modern micro array approaches to identify genes regulated by Pax2. How the chromatin structure of such target genes is affected by the Pax2 PTIP complex will be investigated. |
 ON THE COVER
ON THE COVER
Breast team reviewing a patient's slide. (From left to right) Ghassan Allo, Fellow; Laura Walters, Clinical Lecturer; Celina Kleer, Professor. See Article 2014Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Autopsy Technician draws blood while working in the Wayne County morgue. See Article 2016Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Dr. Sriram Venneti, MD, PhD and Postdoctoral Fellow, Chan Chung, PhD investigate pediatric brain cancer. See Article 2017Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Director of the Neuropathology Fellowship, Dr. Sandra Camelo-Piragua serves on the Patient and Family Advisory Council. 2018Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Residents Ashley Bradt (left) and William Perry work at a multi-headed scope in our new facility. 2019Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Dr. Kristine Konopka (right) instructing residents while using a multi-headed microscope. 2020Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Patient specimens poised for COVID-19 PCR testing. 2021Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Dr. Pantanowitz demonstrates using machine learning in analyzing slides. 2022Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
(Left to Right) Drs. Angela Wu, Laura Lamps, and Maria Westerhoff. 2023Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Illustration representing the various machines and processing used within our labs. 2024Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|

MLabs, established in 1985, functions as a portal to provide pathologists, hospitals. and other reference laboratories access to the faculty, staff and laboratories of the University of Michigan Health System’s Department of Pathology. MLabs is a recognized leader for advanced molecular diagnostic testing, helpful consultants and exceptional customer service.