

 Like prisms spread light in a variety of hues, Dr. Yang Xiao’s talents spread across multiple disciplines, creating a spectrum of color in this world. Dr. Xiao is one of Pathology’s newest faculty members and was recruited as a tenure-track assistant professor. However, she is not only a member of Pathology but also holds joint appointments in Biomedical Engineering and Computational Medicine & Bioinformatics departments and is a member of the Bioscience Initiative Single-Cell Spatial Analysis Program (SCSAP).
Like prisms spread light in a variety of hues, Dr. Yang Xiao’s talents spread across multiple disciplines, creating a spectrum of color in this world. Dr. Xiao is one of Pathology’s newest faculty members and was recruited as a tenure-track assistant professor. However, she is not only a member of Pathology but also holds joint appointments in Biomedical Engineering and Computational Medicine & Bioinformatics departments and is a member of the Bioscience Initiative Single-Cell Spatial Analysis Program (SCSAP).
Xiao was raised in China and completed her undergraduate studies in molecular biology at McGill University in Montreal, Canada. Following graduation, she relocated to New Haven, Connecticut, to pursue a PhD in biomedical engineering at Yale University under the mentorship of Dr. Rong Fan, with a focus on tissue engineering and spatial sequencing hardware innovation. “I investigated how to build a freestanding perfusable vasculature system; how to engineer the cells to perform as vessels and observe how they behave and interact with other brain cells.” Upon completing her PhD, Xiao moved to Columbia University in New York City for her postdoctoral training in biomedical engineering under Dr. Kam Leong. “I applied the spatial omics tools to understand how memory and emotions are encoded in postmortem human brains at the molecular level. I examined how these are organized in neurotypical and in depressed patients’ brains.” After her postdoctoral studies, Xiao came to Michigan in 2025.
“When I was growing up, I wondered how an organism knows where to form limbs and where to form the front and back sides when there is no brain yet? What controls what gets formed, and how do we avoid ending up with too many hands or legs? What is governing the fate of our cells? That information is highly compressed and precisely executed to become an integrated whole. This was a fascinating question to me growing up – how do we know to become human, with our minds, and not a bird or an amphibian?”
Initially, Xiao thought she would become a paleontologist to study the history of animals or evolution. “Later, I realized the technology for us to understand every cell in our bodies is lacking. We often study organisms as collections of independent parts, but in reality, each part is very dependent on the cells around them. This curiosity launched Xiao on her path to pathology, but the decision point came much later, during her PhD program. “One of my committee members, Dr. Alexander Vortmeyer, a neuropathologist, brought me to the pathology room outside the operating room as I was shadowing him. That day changed my life.” Vortmeyer diagnosed three cases while Xiao shadowed him. In one case, a patient’s biopsy did not capture the tumor cells, and the patient was going to have to return in six months for another biopsy. “When I looked at the slide, I saw the hemorrhage, and I could feel the pain of the patient and the weight of uncertainty for the next six months.” Another case involved determining whether a lesion was a low-grade glioma. “In our textbook, we learned that the recommended decision is to proceed with surgery to remove it. However, the patient wanted to wait to be able to attend her grandchild’s graduation ceremony in May. This made me realize that so many factors kick in with medical decisions – it isn’t just what is best for the patient’s health, but also considerations like insurance, finance, lifestyle choices, or special events they want to attend. I switched from bench research to patient-focused science and chose pathology because it translates into making a real difference for patients.”
Xiao’s home in Pathology is within the Experimental Pathology division, working on the bioengineering technology side of molecular pathology. “Single-cell and spatial sequencing focus on molecules and cells with great precision and at a high resolution to understand how cells work together, change their states in response to environmental changes, and save the memory in their epigenome.” Xiao employs spatial technologies, such as Deterministic Barcoding in Tissue for spatial sequencing (DBiT-seq) and Spatial ATAC-seq, which she helped develop during her postdoctoral work, to systematically define the cell states in the human brain.
 Her work in spatial omics is a technological advancement that allows the interrogation of cells or molecules in their original spatial locations. “Modern biology techniques process millions of homogenized cells to collect DNA, RNA, or proteins. The readouts are from one million cells or more, lacking precision of what each single cell does in the disease or development. Spatial sequencing aims to map the original molecule at the twenty picogram level or a 10-micron level to understand cellular function at their original locations.” A picogram is one trillionth of a gram, and a micron is one millionth of a meter.
Her work in spatial omics is a technological advancement that allows the interrogation of cells or molecules in their original spatial locations. “Modern biology techniques process millions of homogenized cells to collect DNA, RNA, or proteins. The readouts are from one million cells or more, lacking precision of what each single cell does in the disease or development. Spatial sequencing aims to map the original molecule at the twenty picogram level or a 10-micron level to understand cellular function at their original locations.” A picogram is one trillionth of a gram, and a micron is one millionth of a meter.
DBiT-seq uses microfluidic systems to deliver DNA oligos as spatial barcodes to cells and conjugate to the poly-adenine tail of messenger RNAs in situ. "Once individual mRNAs are barcoded, we can build sequencing libraries, with up to 2,500 libraries generated from a single tissue slice. and sequence them in bulk." Through sequencing alignment and bioinformatic analysis, the transcriptome’s location is resolved, allowing observations of cell states with high granularity. For example, while bulk RNA or single-cell RNA sequencing may show excitatory neurons, specific neurons function differently based on their neighbors. Their location and microenvironment dictate their behavior. DBiT-seq is a barcoding chemistry that can be extended for labeling RNA, DNA, or surface proteins. “We introduced a sister method named “Patho-DBiT” last year in Cell Journal, demonstrating spatial labeling of tRNA, long non-coding RNA, and microRNA, which opens up RNA biology in both fresh frozen and formalin-fixed paraffin-embedded tissues." Spatial omics integrates spatial transcriptomics, spatial epigenomics, and spatial proteomics. “We can measure all these to understand their specific functions and infer which cells are communicating and how they move in their spatial region.”
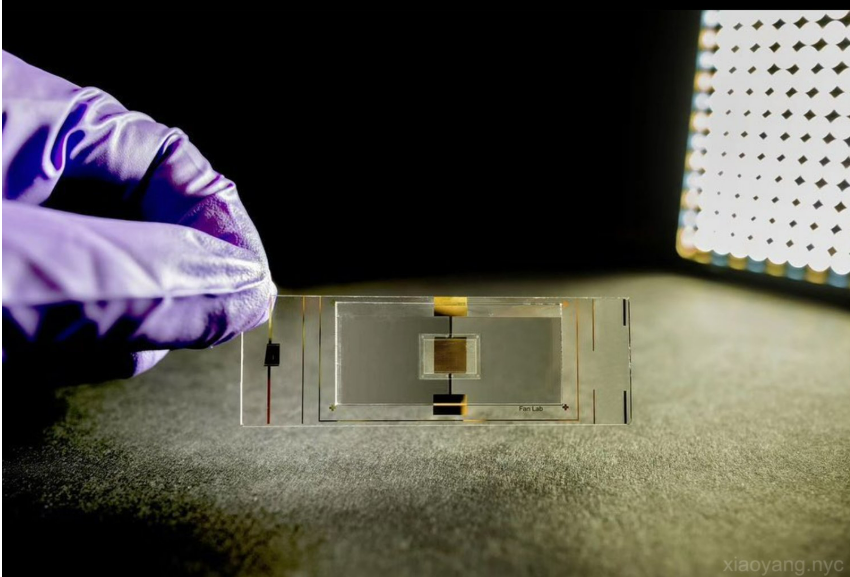 Using this technique, scientists need no prior knowledge of the tissue before analysis. Traditional methods require specific stains for Pathologists to identify certain cells, necessitating some knowledge about the sample. Spatial omics allows for unbiased evaluation. “It is also very high throughput. For example, in the human hippocampus of the brain, we can detect about 20,000 different genes simultaneously. It is a molecular bank where you can call up specific genes to see how they are being expressed and in which part of the cell the expression is happening. The beauty of this is that people can revisit the data and explore deeper. The data is standardized. It is an absolute quantitative measure, eliminating the need for controls required by other methods.” The most significant barrier to utilizing spatial omic methodologies is the expertise needed. An interdisciplinary background is necessary, combining skills in electrical engineering for microfabrication, surface chemistry for barcoding, molecular biology for sequencing library development, and bioinformatics for data analysis. This makes it a high entry barrier for people to enter the spatial omics field.
Using this technique, scientists need no prior knowledge of the tissue before analysis. Traditional methods require specific stains for Pathologists to identify certain cells, necessitating some knowledge about the sample. Spatial omics allows for unbiased evaluation. “It is also very high throughput. For example, in the human hippocampus of the brain, we can detect about 20,000 different genes simultaneously. It is a molecular bank where you can call up specific genes to see how they are being expressed and in which part of the cell the expression is happening. The beauty of this is that people can revisit the data and explore deeper. The data is standardized. It is an absolute quantitative measure, eliminating the need for controls required by other methods.” The most significant barrier to utilizing spatial omic methodologies is the expertise needed. An interdisciplinary background is necessary, combining skills in electrical engineering for microfabrication, surface chemistry for barcoding, molecular biology for sequencing library development, and bioinformatics for data analysis. This makes it a high entry barrier for people to enter the spatial omics field.
The technology for spatial omics was only developed in the last five years, making it a new field. Xiao collaborates with Dr. Joshua Welsh in bioinformatics, utilizing advances in computer vision to integrate and interpretate the spatial data. “Machines may be able to see things that the human eye cannot, such as gene co-expression or transcription factor co-regulation patterns. We are deeply digging into computer science for understanding the gene regulatory network.” In the next five to ten years, Xiao hopes to move this technology from research into patient care and make new biological findings to improve patient care. “My ambition is to apply spatial omics technology in pathology to understand how the brain encodes and stores information such as memories and emotions. Unlike collecting a biopsy or a tumor resection from patients, collecting human brain samples is significantly more challenging.” To overcome this, Xiao is collaborating with Dr. Thomas Sanderson and Dr. Joseph Wider to investigate brain changes following traumatic brain injury, in rodent and swine models. Together, they seek to understand how cells transform in response to major stress events and how chemical changes influence these processes. They are also exploring how axon injuries propagate in spatial contexts. This will require studying the transcriptome and epigenome from brain cells collected at various time points after injury. “The epigenome dictates which region of the DNA is accessible or silenced, leading to differential gene expression across cell types. A cell’s “memory” is encoded in the epigenetic marks, such as histone modifications and chromatin accessibility, enabling the cell to recognize its identity and determine when to function. The knowledge gained in epigenetics is also applicable to understanding how cells function in heart, lung, or other tissues.”
 This type of deep scientific inquiry requires seeing the world differently. Xiao added courses from the Yale School of Art to her portfolio to develop this ability. “I took courses on film photography and darkroom printmaking to train my visual expression. I see myself as a professional large-format photographer, and I’m thriving. This helps me to view spatial data differently, seeing individual cells in the tissues collectively as a landscape and each molecule in the omics as a layer of color shade.” Her photography is beautiful, showing New York City from various perspectives. She also creates portraits of people in the lab and the research community. “I try to capture the most exciting or hardest struggling moments when we are making scientific discoveries or innovations. Illustrating scientists whose work is in progress, telling a story beyond what we publish or what we can do in the lab. It is a story that the general public audience can appreciate. I also want to spotlight the junior trainees who are working in the lab but are not yet being recognized.”
This type of deep scientific inquiry requires seeing the world differently. Xiao added courses from the Yale School of Art to her portfolio to develop this ability. “I took courses on film photography and darkroom printmaking to train my visual expression. I see myself as a professional large-format photographer, and I’m thriving. This helps me to view spatial data differently, seeing individual cells in the tissues collectively as a landscape and each molecule in the omics as a layer of color shade.” Her photography is beautiful, showing New York City from various perspectives. She also creates portraits of people in the lab and the research community. “I try to capture the most exciting or hardest struggling moments when we are making scientific discoveries or innovations. Illustrating scientists whose work is in progress, telling a story beyond what we publish or what we can do in the lab. It is a story that the general public audience can appreciate. I also want to spotlight the junior trainees who are working in the lab but are not yet being recognized.”
“My art ties back into my science. I am a visual person, and spatial data is visual. We not only use CPUs to compute, but also GPUs. The graphical models, instead of the logical computations, recognize the patterns and dependencies in the spatial data. My art contributes to how I visualize my science.”
Please be sure to welcome Dr. Xiao to the Pathology Department and take a few moments to see her fascinating artwork.