
By Rosemary Clandos | labblog.uofmhealth.org | 5 April
 Dr. Thomas Giordano / Photo: Dustin Johnston.Thirteen years after its initial funding, the Cancer Genome Atlas (TCGA) has analyzed 2.5 petabytes of data representing 33 different types of cancer.
Dr. Thomas Giordano / Photo: Dustin Johnston.Thirteen years after its initial funding, the Cancer Genome Atlas (TCGA) has analyzed 2.5 petabytes of data representing 33 different types of cancer.
That’s no small feat: One petabyte has the storage capacity of 212,000 DVDs.
The project has created thorough, multi-dimensional maps of the key genomic changes in both common and rare cancers.
Now, a series of 25 papers publishing across the Cell Press family of journals summarizes its final project, PanCancerAtlas, the most comprehensive cross-cancer genomic analysis available.
“Nearly all TCGA projects identified previously unrecognized molecular subgroups and, moreover, genomic insights from some projects (e.g. thyroid cancer) have catalyzed significant revisions to how we categorize some cancers,” says Thomas Giordano, MD, PhD, Henry Clay Bryant professor of pathology at Michigan Medicine. Giordano has served as a member of the PanCancerAtlas steering committee, and he co-led the projects on thyroid and adrenal cancer.
The University of Michigan has contributed tumor samples and led several of the TCGA projects. Participating faculty include David Beer, PhD, who contributed to the esophageal cancer project, and Thomas Carey, PhD, who contributed to the head and neck squamous cell carcinoma project.
Giordano discussed the project and its implications:
What were the goals for TCGA and what can we now take away from it?
Giordano: For the 10-year project, there were three primary goals: to better understand the molecular underpinnings of cancer, to derive original molecular taxonomies, and to identify novel molecular therapeutic targets.
We learned a great deal about the molecular basis of cancer and discovered new mutations, which helped improve molecular diagnostics. The research helped us understand the functional consequences of specific mutations, particularly, how they drive the biology of tumors. Nearly all projects identified three to five molecular subgroups of cancer, some of which had not been previously recognized.
For example, in papillary thyroid cancer, two mutations involving proteins in the same signaling pathway were believed to be essentially functionally equivalent. But our data revealed different signaling properties. Understanding this impacts tumor classification, which affects patient care and treatment.
As a pathologist, it’s been exciting to understand the underlying genomics and correlate the findings with the microscopic appearance of tumors.
With the thyroid cancer project, we were concerned that the project might be uninteresting because it was focused on the most indolent type of thyroid cancer and excluded the most clinically aggressive types. However, we learned so much about thyroid cancer from that project and it provided a foundation for comparison with other projects.
Now, when researchers study an aggressive type of thyroid cancer, they can compare their results to TCGA and derive increased understanding by that comparison. This holds true for other types of cancer as well.
Can you talk about the size and scope of the project?
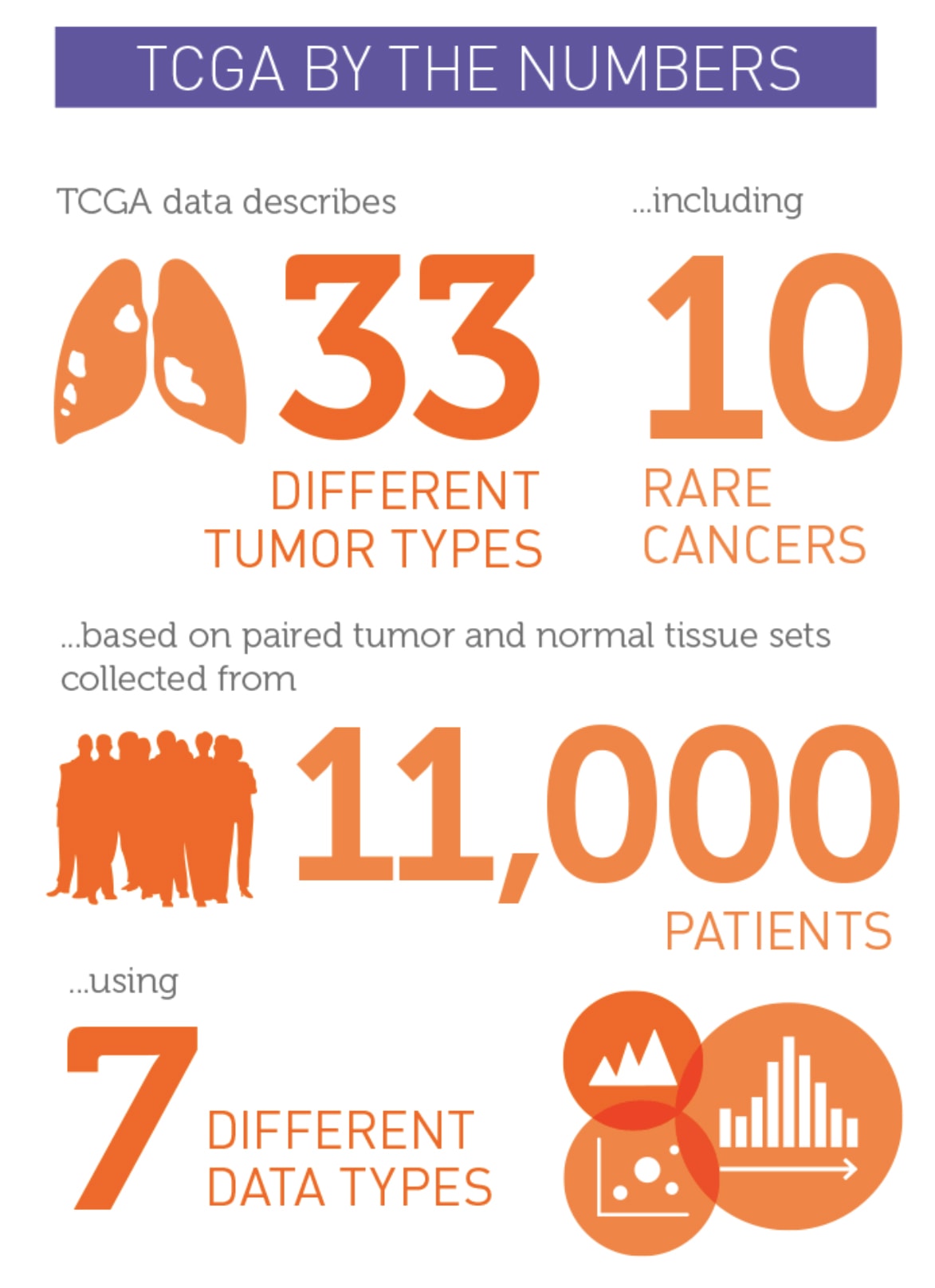 Giordano: TCGA built a standardized platform for the genomic characterization of cancer. Thirty-three cancer types were examined; 23 were common cancers and 10 were rare. A total of 11,000 tumor types were studied.
Giordano: TCGA built a standardized platform for the genomic characterization of cancer. Thirty-three cancer types were examined; 23 were common cancers and 10 were rare. A total of 11,000 tumor types were studied.
One of the rare cancer projects was on adrenocortical cancer. The TCGA project was hesitant to address adrenal cancer because it is so rare, but my colleague Gary Hammer, MD, PhD, director of the endocrine oncology program at U-M, and I were instrumental in making that happen. We assembled an international collaboration to study enough tumors.
We were able to identify three prognostic subgroups, which is a true advance for the field. Now we’re trying to translate those findings to the clinic and tailor patient care to the biology of these tumors.
How did the discoveries impact the classification of tumors?
Giordano: It changes the way pathologists think about cancer and re-examines the proper classification of these tumors. When tumor morphology was integrated with genomic features and molecular subtypes, knowledge increased. Insights about genomics for thyroid cancers and other cancers have catalyzed or accelerated revisions of tumor taxonomies, or how we classify tumors.
What problems were uncovered by the new cancer taxonomy?
Giordano: Tumors are routinely classified based on what pathologists see through the microscope. However, two tumors that look similar might have different underlying genetic abnormalities, which can only be revealed by molecular pathology. Fully understanding the genetic landscape in a given tumor type advances molecular pathology testing and informs therapeutic decisions. Instead of treating people with non-specific chemotherapies, we are undergoing a paradigm shift to using molecularly targeted therapies.
This will fundamentally change how patients are treated. As we move toward more precision therapy, some patients with specific genetic alterations will respond to drugs. Therapy that works on one mutation won’t work on another.
What are some of the challenges moving forward?
Giordano: The main challenge is that genomic characterization of cancer is outpacing development of targeted therapeutics. Some frequently mutated genes are still difficult to target. So we may know the alterations, but we haven’t found drugs to block the alterations.
A number of pathways are so difficult to target that some pharmaceutical companies have de-empathized drug development for those pathways.
What were some of the limitations of the research?
Giordano: TCGA didn’t do research on all types of cancer, but the research will be used for comparison and discoveries for many years.
How might these findings affect future research?
 Photo: Dustin Johnston.Giordano: There’s a paradigm shift underway and it relates to the transition to precision pathology. For example, you don’t treat lung cancer with traditional chemotherapy until you’ve determined if there are genetic alterations that can be targeted.
Photo: Dustin Johnston.Giordano: There’s a paradigm shift underway and it relates to the transition to precision pathology. For example, you don’t treat lung cancer with traditional chemotherapy until you’ve determined if there are genetic alterations that can be targeted.
This will be more work for pathologists, but it is exciting work. We will continue to do microscopy, but we’ll be doing more and more molecular pathology as we look for actionable genetic alterations.
In the future, results from genomic characterization of tumors will be combined with big data analytics to help guide therapy. Pathologists will be using image analysis and big data combined with insights derived from systems biology and artificial intelligence.
 ON THE COVER
ON THE COVER
Breast team reviewing a patient's slide. (From left to right) Ghassan Allo, Fellow; Laura Walters, Clinical Lecturer; Celina Kleer, Professor. See Article 2014Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Autopsy Technician draws blood while working in the Wayne County morgue. See Article 2016Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Dr. Sriram Venneti, MD, PhD and Postdoctoral Fellow, Chan Chung, PhD investigate pediatric brain cancer. See Article 2017Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Director of the Neuropathology Fellowship, Dr. Sandra Camelo-Piragua serves on the Patient and Family Advisory Council. 2018Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Residents Ashley Bradt (left) and William Perry work at a multi-headed scope in our new facility. 2019Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Dr. Kristine Konopka (right) instructing residents while using a multi-headed microscope. 2020Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Patient specimens poised for COVID-19 PCR testing. 2021Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Dr. Pantanowitz demonstrates using machine learning in analyzing slides. 2022Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
(Left to Right) Drs. Angela Wu, Laura Lamps, and Maria Westerhoff. 2023Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|
 ON THE COVER
ON THE COVER
Illustration representing the various machines and processing used within our labs. 2024Department Chair |

newsletter
INSIDE PATHOLOGYAbout Our NewsletterInside Pathology is an newsletter published by the Chairman's Office to bring news and updates from inside the department's research and to become familiar with those leading it. It is our hope that those who read it will enjoy hearing about those new and familiar, and perhaps help in furthering our research. CONTENTS
|

MLabs, established in 1985, functions as a portal to provide pathologists, hospitals. and other reference laboratories access to the faculty, staff and laboratories of the University of Michigan Health System’s Department of Pathology. MLabs is a recognized leader for advanced molecular diagnostic testing, helpful consultants and exceptional customer service.